There are now over 50 human peptides and proteins associated with amyloid disease and the formation of insoluble, fibrillar deposits. Interestingly, these proteins share little in common – they have variable native topologies and secondary structure content (some are even natively disordered), amino acid composition, and native functions. Despite these differences, the resulting mature fibrils and intermediate species share many characteristics. Traditionally, the formulation of diagnostic and/or therapeutic strategies requires structural insight into the target protein. However, the heterogeneous – and potentially disordered – ensemble of intermediate amyloid species largely preclude structural insight through traditional methods such as X-ray crystallography or NMR spectroscopy.
To fill this gap, we perform atomistic simulations of amyloid proteins under amyloidogenic conditions to model the early conformational changes that occur during amyloidogenesis. In simulations of multiple proteins, including transthyretin, superoxide dismutase, prion, and lysozyme – we have observed the formation of an unusual type of secondary structure that we have dubbed α-sheet. The unique properties of this secondary structure have guided our lab’s central hypothesis regarding amyloidogenesis and led to the development of peptides capable of inhibiting amyloid formation in multiple systems.
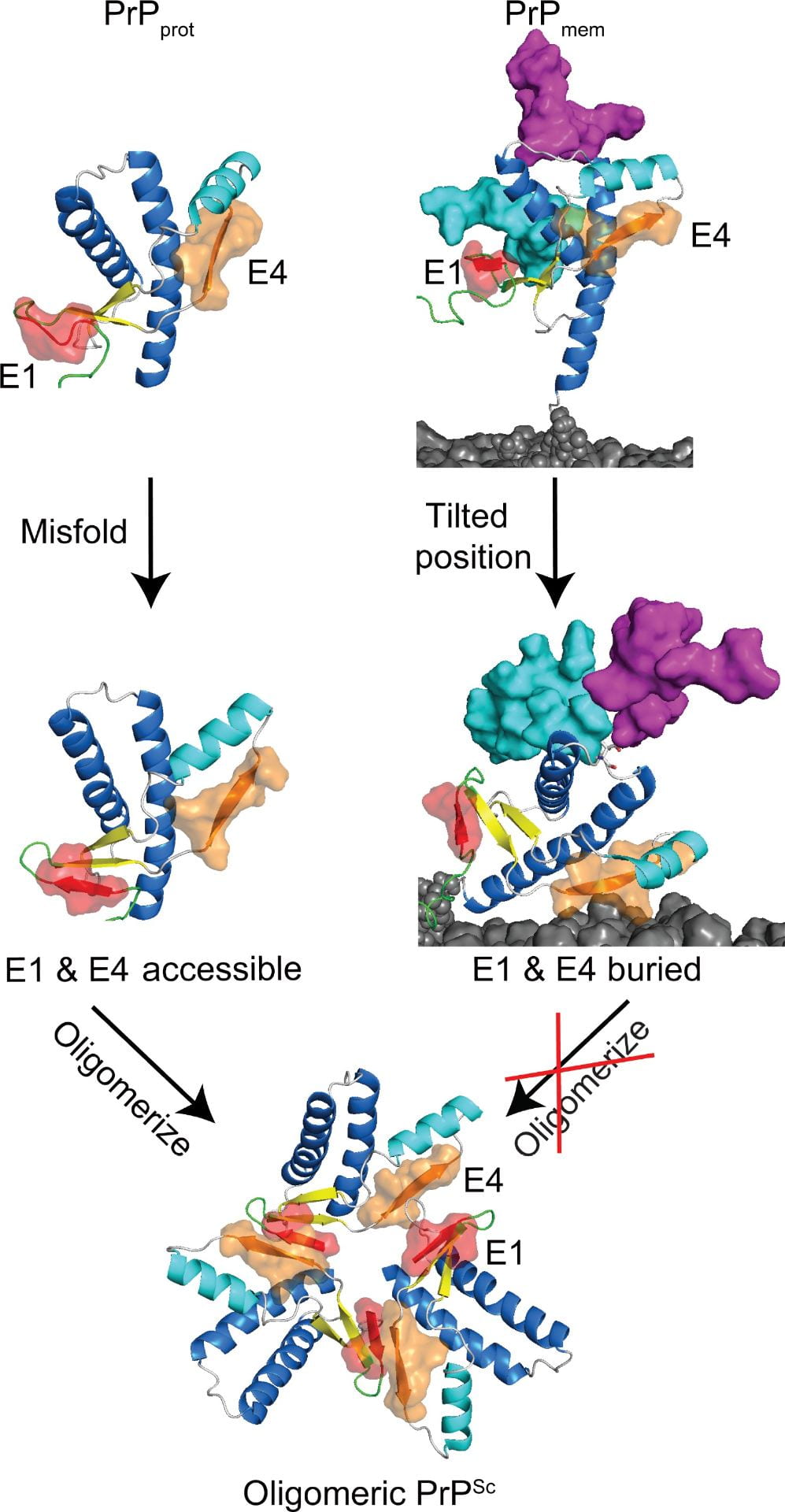
(Left) Schematic of prion protein (PrP) conversion. Without glycans and the membrane environment, PrPprot misfolds and both E1 and E4 strands are accessible, allowing for oligomerization. As for PrPmem, the oligomerization sites E1 and E4 are buried or near the membrane surface, which would interfere of PrPSc to PrPC and propagation of PrPSc. (image source)