Optical setup and modifications of Bruker amaZon mass spectrometer for tandem MS and UV/Vis photodissociation action spectroscopy, tuning from 210 to 700 nm. The tunable setup incorporated a fast steering mirror to improve beam alignment and UVPD reproducibility. (A. Dang et. al, J. Am. Soc. Mass Spectrom. 2019, 30, 1558-1564).
Gas Phase DNA Radicals Characterized by UV-Vis Photodissociation Action Spectroscopy and DFT Calculations
Although DNA is chemically very stable, it is susceptible to chemical transformations as a result of ionization that are often summarized under the the term of DNA damage. DNA damage is of concern because it temporarily or permanently disrupts the genetic information stored for the lifetime of the individual.
One of the two major mechanism of ionizing DNA damage is the direct ionization with an energetic particle to produce cation radicals. These radical intermediates can undergo further reactions with environment to produce modified nucleobases, undergo nucleobase loss or strand breakage. The second major mechanism is the low-energy electron capture by the nucleobases, forming transient anion radicals that can spontaneously eliminate the affected nucleobase of protonated by solvent to form radical hydrogen atom adducts. Despite decades of studies, these radical intermediates of DNA ionization have not been characterized spectroscopically. The main difficulty in studying nucleobase and nucleoside cation radicals in solution is the very rapid proton transfer onto the solvent.
We apply the methods of gas-phase ion chemistry to generate DNA and RNA-relatived cation radicals and investigate these transient radical intermediates via UVPD action spectroscopy combined with DFT calculations.
We’ve investigated cation radicals of DNA nucleobases, di- and tetranucleotides. We have plans to investigate new nucleobase systems, as well as new synthetic methods to generate these radical species.
This figure shows the conformational collapse of the dinucleotide system (dGG+2H)2+ upon one-electron reduction and its corresponding UVPD action spectrum. (Reproduced from Y. Liu et. al, J. Phys. Chem. B, 2018, 122, 9665-9680.)
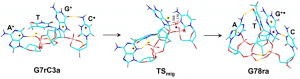
This figures shows the isomerization and its transition state of a tetranucleotide system, dGATC, as a hydrogen migrates from the cytosine to the guanine ring. (Reproduced from S.R. Huang et. al, J. Phys. Chem. B, 2020, submitted.)
Photodissociative Cross-Linking of Peptide-Peptide, Peptide-DNA, Peptide-Small Molecule Non-covalent Complexes in the Gas-Phase
Non-covalent interactions underlie most biological structures, reactions, signaling, transport, etc. Determination of structures of non-covalent complexes primarily relies on condense phase methods, such as X-ray crystallography, NMR, and cyrogenic electron microscopy. Our lab, with our experience in tandem MS, has introduced gas-phase photodissociative cross-linking to investigate non-covalent complexes. We utilize the chemistry of the diazirine photo-chemical tag, where the photolytically generated carbene reactive intermediate inserts itself into H-X bonds to form new C-H and C-X bonds within the diazirine-tagged ion-molecule complex. The cross-linked covalent products are analyzed by gas-phase sequencing to identify cross-link positions. In addition, Born-Oppenheimer Molecular Dynamics are used to evaluate positions of close-contact between the photolabel and atoms in the target molecule or ion.
We have developed new tagging protocols and applied it to peptide-peptide complexes, amyloid fragments, phosphopeptide-arginine pairs, and peptide-DNA complexes. We plan to explore and develop a new nitrene-based photo-chemical tag to expand our investigation of peptide-DNA, peptide-small molecule and peptide-sugar non-covalent complexes.
This figure shows three thermodynamically-favored DFT optimized structures of the peptide-peptide non-covalent complex (C*AQK+LLSPGH+H)+. Atom color coding are: cyan/magenta = C, blue = N, red = O, yellow = S, gray = H; the diazirine chemical tag is annotated with asterisks. In addition, only exchangeable hydrogens are displayed. (Reproduced from Y. Liu et. al, Chem. Eur. J. 2018, 24, 9259-9263.)
This figure shows some of the DFT optimized strucutres of the zwitterionic non-covalent peptide-DNA complex (C*AQK+dGG+H)+. Atom color codes are: light green = peptide C’s , cyan = DNA C’s, red = O, blue = N, yellow = S, gray = H; the diazirine chemical tag is annotated with asterisks. In addition, only exchangeable hydrogens are displayed. (Reproduced from Y. Liu et. al, J. Am. Soc. Mass Spectrom. 2019, 30, 1992-2006.)