Publications
Justin Kollman’s Google Scholar Profile
| A family of bacterial actin homologues forms a 3-stranded tubular structure Bergeron J, Kollman JM 2023 prepring:bioarxiv |
 |
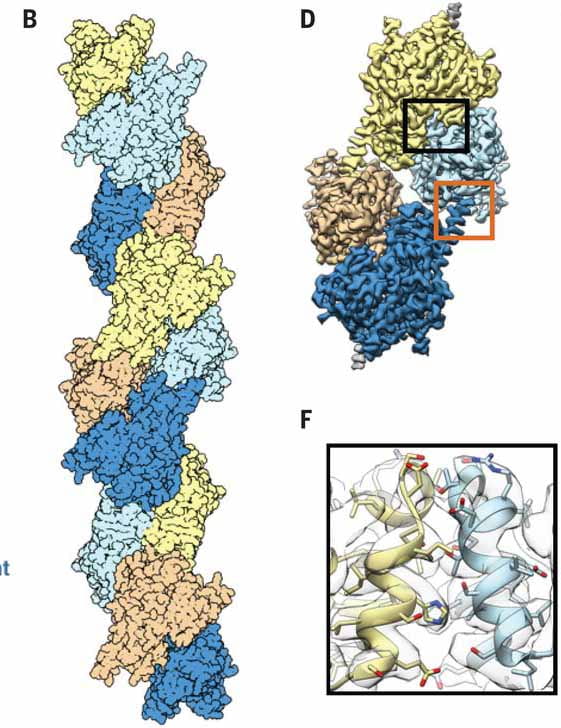
| De novo design of pH-responsive self-assembling helical protein filaments Shen H, Lynch EM, Akkineni S, Watson JL, Decarreau J, Bethel NP, Benna I, Sheffler W, Farrell D, DiMiaio F, Derivery E, De Yoreo JJ, Kollman JM, Baker D Nature Nanotechnology 2024 |
 |
| Blueprinting expandable nanomaterials with standardized protein building blocks Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Borst AJ, Han HL, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Coventry B, Brunette TJ, Liu Y, Dauparas J, Ekiert D, Kollman JM, Bhabha G, Baker D Nature 2023 preprint: bioarxiv |
 |
| Light-sensitive phosphorylation regulates enzyme activity and filament assembly of human IMPDH1 retinal splice variants Calise SJ, O’Neill AG, Burrell AL, Dickinson MS, Molfino J, Clarke C, Quispe J, Sokolov D, Buey RM, Kollman JM Journal of Cell Biology 2023 preprint: bioarxiv |
 |
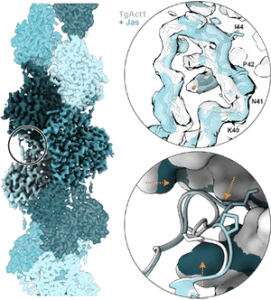
| Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover Hvorecny KL, Sladewski TE, De La Cruz EM, Kollman JM, Heaslip AT Nature Communications 2023 preprint: bioarxiv |
 |
| Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation O’Neill AG, Burrell AL, Zech M, Elpeleg O, Harel T, Edvardson S, Shaked HM, Rippert AL, Nomakuchi T, Izumi K, Kollman JM Journal of Biological Chemistry 2023 preprint: bioarxiv |
 |
| Human PRPS1 filaments stabilize allosteric sites to regulate activity Hvorecny KL, Hargett KA, Quispe J, Kollman JM Nature Structural & Molecular Biology 2023 preprint: bioarxiv |
 |
| Greater than the sum of parts: Mechanisms of metabolic regulation by enzyme filaments Hvorecny KL, Kollman JM Current Opinion in Structural Biology 2023 |
 |
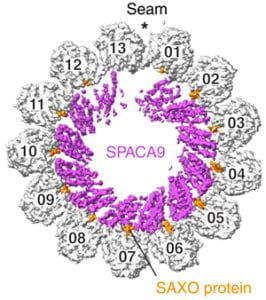
| SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules Gui M, Croft JT, Zabeo D, Acharya V, Kollman JM, Burgoyne T, Höög JL, Brown A Proceedings of the National Academy of Sciences USA 2022 |
 |
| The Giardia ventrolateral flange is a lamellar membrane protrusion that supports attachment Hardin WR, Alas GCM, Taparia N, Thomas EB, Steele-Ogus M, Hvorecny KL, Halpern AR, Tůmová P, Kollman JM, Vaughan JC, Sniadecki NJ, Paredez AR PLOS Pathogens 2022 preprint: bioRxiv |
 |
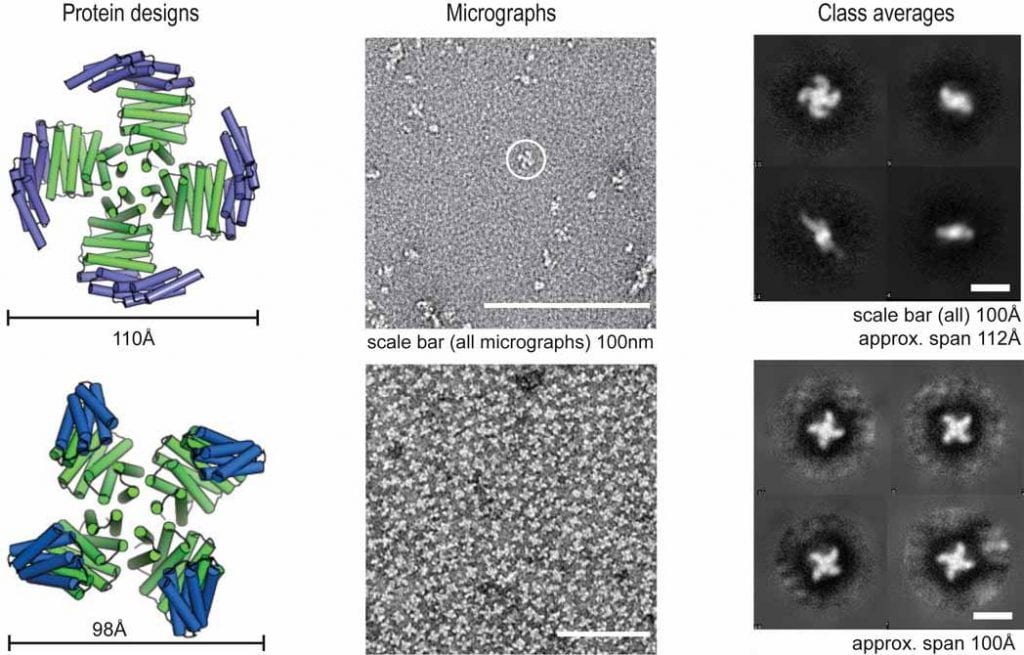
| Computational design of mechanically coupled axle-rotor protein assemblies Courbet A, Hansen JM, Hsia Y, Bethel N, Park Y, Xu C, Moyer A, Boyken S, Ueda G, Natterman U, Nagarajan D, Silva D, Sheffler W, Quispe J, King N, Bradley P, Kollman JM, Baker D Science 2022 preprint: bioRxiv |
 |
| IMPDH dysregulation in disease: a mini review Burrell AL, Kollman JM Biochemical Society Transactions 2022 |
 |
| IMPDH1 retinal variants control filament architecture to tune allosteric regulation Burrell AL, Nie C, Said M, Simonet JC, Fernández-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM Nature Structural & Molecular Biology 2022 preprint: bioRxiv |
 |
| A highly conserved zebrafish IMPDH retinal isoform produces the majority of guanine and forms dynamic protein filaments in photoreceptor cells Cleghorn WM, Burrell AL, Giarmarco MM, Brock DC, Wang Y, Chambers ZS, Du J, Kollman JM, Brockerhoff SE Journal of Biological Chemistry 2021 |
 |
| Cryo-EM Structures of CTP Synthase Filaments Reveal Mechanism of pH-Sensitive Assembly During Budding Yeast Starvation Hansen JM, Horowitz A, Lynch EM, Farrell DP, Quispe J, DiMaio F, Kollman JM eLife 2021 preprint: bioRxiv |
 |
| Structural basis for isoform-specific inhibition of human CTPS1 Lynch EM, DiMattia MA, Albanese S, van Zundert GCP, Hansen JM, Quispe JD, Kennedy MA, Verras A, Borrelli K, Toms AV, Kaila N, Kreutter KD, McElwee JJ, Kollman JM Proceedings of the National Academy of Sciences USA 2021 |
 |
| Selective activation of PFKL suppresses the phagocytic oxidative burst Amara N, Cooper MP, Voronkova MA, Webb BA, Lynch EM, Kollman M, Ma T, Yu K, Lai Z, Sangaraju D, Kayagaki N, Newton K, Bogyo M, Staben ST, Dixit VM Cell 2021 |
 |
| Mulitmeric antibodies from antigen-specific human IgM+ memory B cells restrict plasmodium parasites Thouvenel CD, Fontana MC, Netland J, Krishnamurty AT, Takehara K, Chen Y, Singh S, Miura K, Keitnay GJ, Lynch EM, Portugal S, Miranda MC, King NP, Kollman JM, Crompton PD, Long C, Pancera M, Rawlings DJ, Pepper M Journal of Experimental Medicine 2021 |
 |
| BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1 Witus SR, Burreell AL, Farrell DP, Kang J, Wang M, Hansen JM, Pravat A, Tuttle LM, Steward MD, Brzovic PS, Chatterjee C, Zhao W, DiMaio F, Kollman JM, Klevit RE Nature Structural and Molecular Biology 2021 |
 |
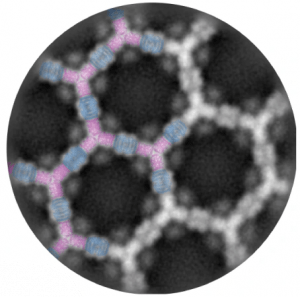
| Design of biologically active binary protein 2D materials. Ben-Sasson AJ, Watson J, Sheffler W, Johnson MC, Bittleston A, Logeshwaran S, Decarreau J, Jiao F, Chen J, Mela I, Drabek AA, Jarrett SM, Blacklow SC, Kaminski CF, Hura GL, De Yoreo JJ, Kollman JM, Ruohola-Baker H, Delivery E, Baker D Nature 2021 preprint: bioRxiv |
 |
| Filament formation by metabolic enzymes-A new twist on regulation Lynch EM, Kollman JM, Webb BA Current Opinion in Cell Biology 2020 |
 |
| Computational design of transmembrane pores. Xu C, Lu P, Gamal TM, Pei X-Y, Johnson MC, Uyeda A, Bick MJ, Xu Q, Jiang D, Bai H, Reggiano G, Yang H, Brunette TJ, Dou J, Ma D, Lynch EM, Boyken SE, Huang PS, Stewart L, DiMaio F, Kollman JM, Luisi BF, Matsuura T, Catterall WA, Baker D Nature 2020 |
 |
| CTP synthase polymerization in germline cells of the developing Drosophila egg supports fecundity under nutrient stress. Simonet JC, Foster MJ, Lynch EM, Kollman JM, Nicholas E, O’Reilly AM, Peterson JR Biology Open 2020 |
 |
| Lysogenic symbiosis in SAR11. Morris RM, Cain KR, Hvorecny KL, Kollman JM Nature Microbiology 2020 |
 |
| Freedom of assembly: Metabolic enzymes come together. Simonet JC, Burrell AL, Kollman JM, Peterson JR Molecular Biology of the Cell 2020 |
 |
| Independent evolution of polymerization in the Actin ATPase clan regulates hexokinase activity. Stoddard PR, Lynch EM, Farrell DP, Dosey AM, William TA, Kollman JM, Murray AW, Garner EC Science 2020 preprint: bioRxiv |
 |
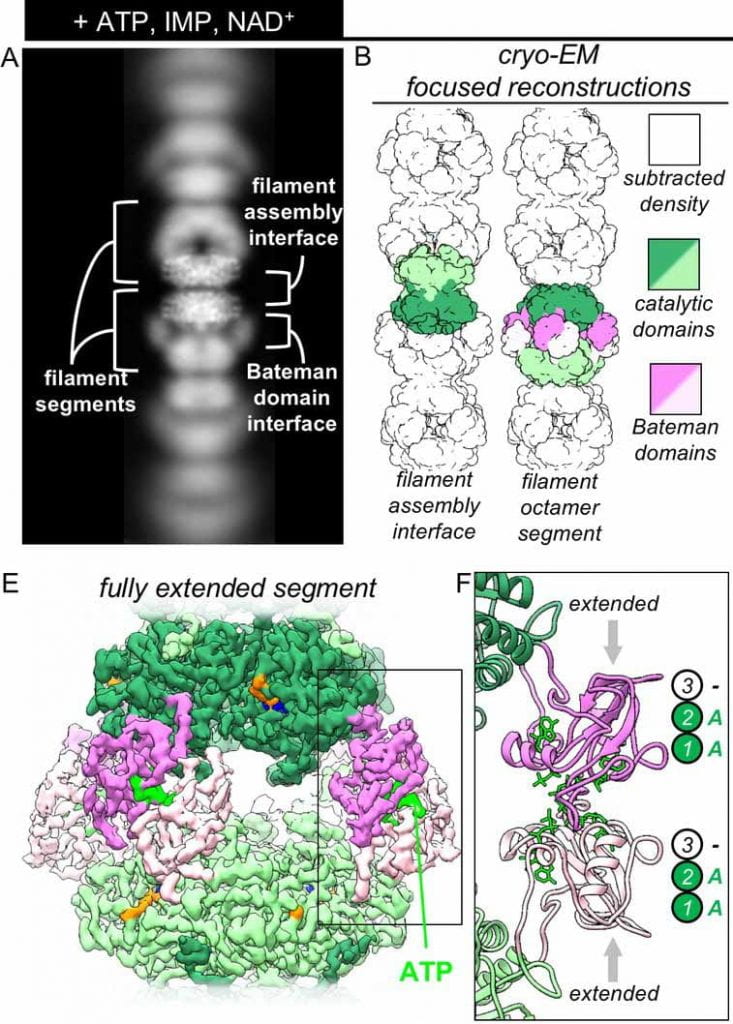
| Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. Johnson MC, Kollman JM eLife 2020preprint: bioaRxiv |
 |
| Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Lynch EM, Kollman JM Nature Structural & Molecular Biology 2020 preprint: bioaRxiv |
 |
| Modular repeat protein sculpting using rigid helical junctions. Brunette TJ, Bick MJ, Hansen JM, Kollman JM, Baker D Proceedings of the National Academy of Sciences USA 2020 |
 |
| A mechanistic understanding of nonclassical hydrothermally synthesized sodium yttrium fluoride nanowires. Bard BA, Zhou X, Xia X, Zhu G, Lim MB, Kim SM, Johnson MC, Kollman JM, Marcus MA, Spurgeon SP, Chun J, Perea DE, Devaraj A, De Yoreo JJ, Pauzauskie P Chemistry of Materials 2020 |
 |
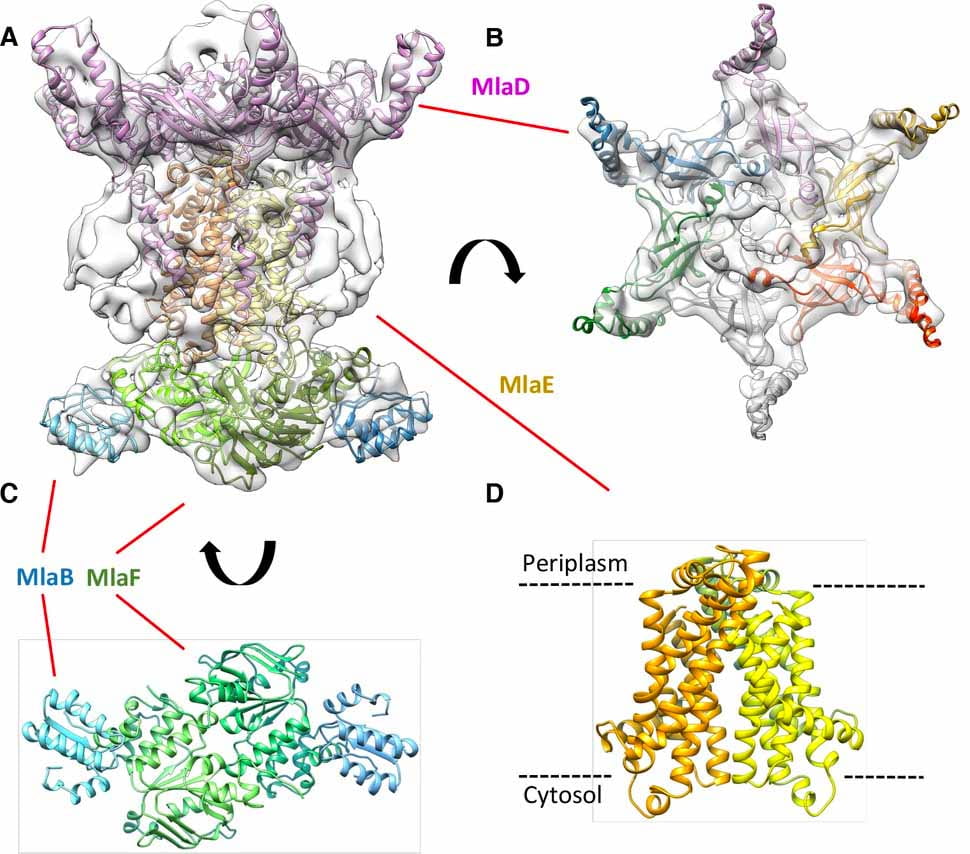
| The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, Kollman JM, Miller SI eLife 2019 preprint: bioRxiv |
 |
| Self-assembling 2D arrays with de novo protein building blocks. Chen Z, Johnson MC, Chen J, Bick MJ, Boyken SE, Lin B, De Yoreo JJ, Kollman JM, Baker D, DiMaio F Journal of the American Chemical Society 2019 |
 |
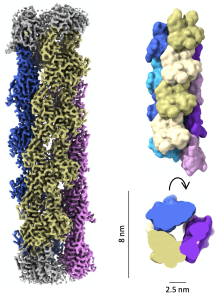
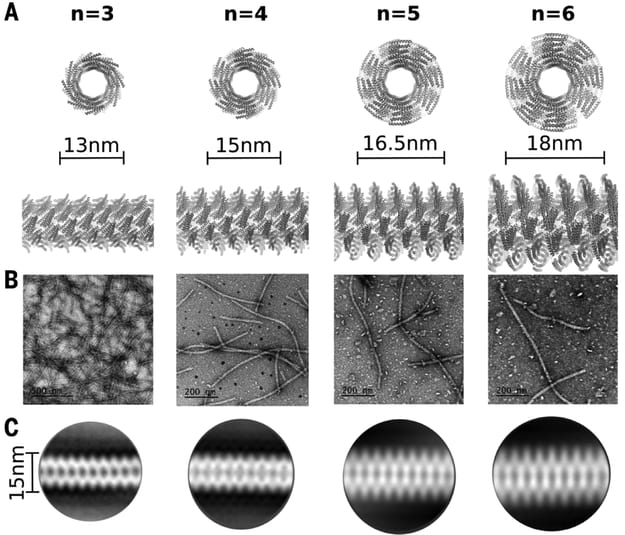
| De novo design of self-assembling helical protein filaments. Shen H, Fallas JA, Lynch E, Sheffler W, Parry B, Jannetty N, Decarreau J, Wagenbach M, Chen J, Wang L, Dowling Q, Oberdorfer G, De Yoreo J, Wordeman L, Jacobs-Wagner C, Kollman JM, Baker D. (2018) Science 2018 |
 |
| The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth. Chaban S, Jariwala S, Hsu C-T, Redemann S, Kollman JM, Müller-Reicher T, Sept D, Bui KH, Brouhard GJ Developmental Cell 2018 |
 |
| T Cell Activation Triggers Reversible Inosine-5′-Monophosphate Dehydrogenase Assembly. Duong-Ly KC, Kuo Y-M, Johnson MC, Kollman JM, Soboloff J, Rall GF, Andrews A, Peterson J. Journal of Cell Science 2018 preprint: bioRxiv |
 |
| Radiant star nanoparticle prodrugs for the treatment of intracellular alveolar infections. Das D, Srinivasan S, Brown FD, Su FY, Burrell AL, Kollman JM, Postma A, Ratner DM, Stayton PS, Convertine AJ Polymer Chemistry 2018 |
 |
| Cryo-EM structure of the bacterial actin AlfA reveals unique assembly and ATP-binding interactions and the absence of a conserved subdomain. Usluer G, Dimaio F, Yang S, Hansen JM, Polka JK, Mullins RD, Kollman JM Proceedings of the National Academy of Sciences USA 2018 preprint: bioRxiv |
 |
| The tetrameric kinesin Kif25 suppresses pre-mitotic centrosome separation to establish proper spindle orientation. Decarreau J, Wagenbach M, Lynch E, Halpern AR, Vaughn JC, Kollman JM, Wordeman L Nature Cell Biology 2017 |
 |
| Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Bergeron JR, Hutto R, Ozyamak E, Hom N, Hansen J, Draper O, Byrne ME, Keyhani S, Komeili A, Kollman JM Protein Science 2017 |
 |
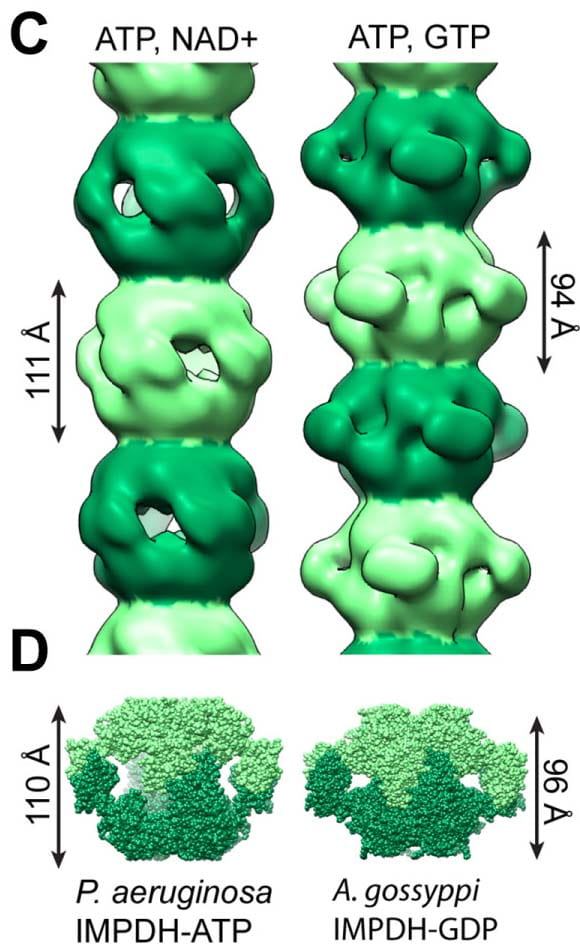
| Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Anthony S, Burrell AL, Johnson MC, Duong-Ly K, Kuo Y-M, Michener P, Andrews A, Kollman JM, Peterson JR Molecular Biology of the Cell 2017 |
 |
| Human CTP synthase filament structure reveals the active enzyme conformation. Lynch EM, Hicks DR, Shepherd M, Endrizzi JA, Maker A, Barry RM, Gitai Z, Baldwin EP, Kollman JM Nature Structural & Molecular Biology 2017 |
 |
| The glycolytic enzyme Phosphofructokinase-1 assembles into filaments. Webb BA, Dosey AM, Wittman T, Kollman JM, Barber DL. Journal of Cell Biology 2017 |
 |
| Structure of UDP-glucose:glycoprotein glucosyltransferase, a sensor of misfolded glycoproteins, by single particle electron microscopy. Calles-Garcia D, Yang M, Soya N, Melero R, Ménade M, Ito Y, Vargas J, Lukacs GL, Kollman JM, Kozlov G, Gehring K. Journal of Biological Chemistry 2017 |
 |
| Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. SnijderJ, Borst AJ, Dosey A, Walls AC, Burrell A, Reddy VS, Kollman JM, Veesler D Journal of Structural Biology 2017 |
 |
| Inhibition of E. coli CTP synthetase by NADH and other nicotinamides, and their synergy with CTP and GTP. Habrian C, Chandrasekhara A, Shahrvini B, Hua B, Lee J, Jesinghaus J, Barry R, Gitai Z, Kollman JM, Baldwin EP Biochemistry 2016 |
 |
| Structure of γ-tubulin small complex based on a cryo-EM map, chemical cross-links, and a remotely related structure. Greenberg CH, Kollman JM, Zelter A, Johnson R, MacCoss MJ, Davis TN, Agard DA, Sali A Journal of Structural Biology 2016 |
 |
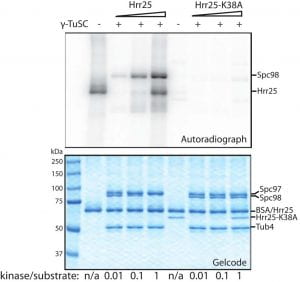
| Interaction of CK1δ with γTuSC ensures proper microtubule assembly and spindle positioning. Peng Y, Moritz M, Han X, Giddings TH, Lyon A, Kollman JM, Winey M, Yates J, Agard DA, Drubin DG, Barnes G Molecular Biology of the Cell 2015 |
 |
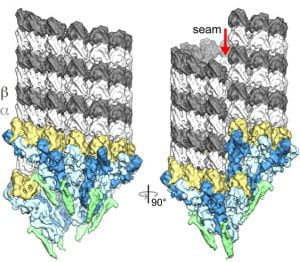
| Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez J-J, Sali A, Kilmartin J, Davis TN, Agard DA Nature Structural and Molecular Biology 2015 |
 |
| Large-scale filament formation inhibits the activity of CTP synthase. Barry RM, Bitbol AF, Lorestani A, Charles EJ, Habrian CH, Hansen JM, Li HJ, Baldwin EP, Wingreen NS, Kollman JM, Gitai Z eLife 2014 |
 |
| Bacterial actins and their diversity. Ozyamak E, Kollman JM, Komeili A Biochemistry 2013 |
 |
| Accessory factors promote AlfA-dependent plasmid segregation by regulating filament nucleation, disassembly, and bundling. Polka JK, Kollman JM, Mullins RD Proceedings of the National Academy of Sciences USA 2013 |
 |
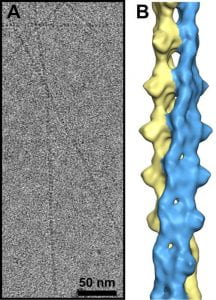
| The bacterial actin MamK: in vitro assembly behavior and filament architecture. Ozyamak E, Kollman JM, Agard DA, Komeili A Journal of Biological Chemistry 2013 |
 |
| Microtubule nucleation by γ-tubulin complexes. Kollman, JM, Merdes, A, Mourey, L, Agard, DA Nature Reviews Molecular Cell Biology 2011 |
 |
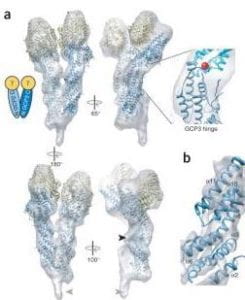
| Insight into γ-tubulin complex assembly from the crystal structure of GCP4. Guillet V, Knibiehler M, Gregory-Pauron L, Remy M-H, Cemin C, Raynaud-Messina B, Bon C, Kollman JM, Agard DA, Mourey L, Merdes A Nature Structural & Molecular Biology 2011 |
 |
| Architecture and assembly of a divergent member of the ParM family of bacterial actin like proteins. Rivera, CR, Kollman, JM, Polka, JK, Agard, DA, Mullins, RD Journal of Biological Chemistry 2011 |
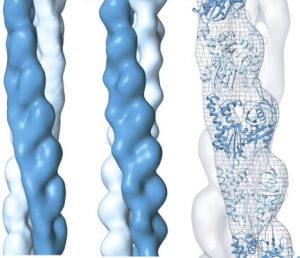 |
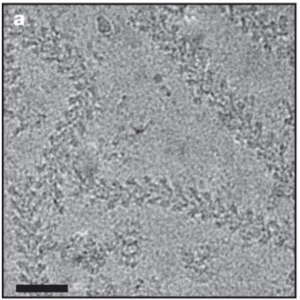
| Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Kollman, JM, Polka, JK, Zelter, A, Davis, TN, Agard, DA Nature 2010 |
 |
| Localization and orientation of the γ-Tubulin Small Complex components using protein tags as labels for single particle EM. Choy, RM, Kollman, JM, Zelter, A, Davis, TN, Agard, DA Journal of Structural Biology 2009 |
 |
| The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. Polka, JK, Kollman, JM, Agard, DA, Mullins RD Journal of Bacteriology 2009 |
 |
| Two families of Synthetic peptides that enhance fibrin turbidity and delay fibrinolysis by different mechanisms. Pandi L, Kollman JM, Lopez-Lira F, Burrows J, Riley M, Doolittle RF Biochemistry 2009 |
 |
| Crystal structure of human fibrinogen. Kollman JM, Pandi L, Sawaya MR, Riley M, Doolittle RF Biochemistry 2009 |
 |
| The structure of the γ-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Kollman JM, Zelter A, Muller EG, Fox B, Davis TN, Rice LM, Agard DA Molecular Biology of the Cell 2008 |
 |
| Automated acquisition of electron microscopic random conical tilt sets. Zheng SQ, Kollman JM, Braunfeld MB, Sedat JW, Agard DA Journal of Structural Biology 2006 |
 |
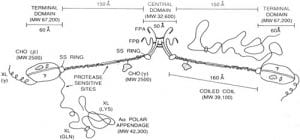
| Natively unfolded regions of the vertebrate fibrinogen molecule. Doolittle RF, Kollman JM Proteins 2006 |
 |
| The 17 Å structure of the 420 kDa lobster clottable protein by single particle reconstruction from cryoelectron micrographs. Kollman JM, Quispe J Journal of Structural Biology 2005 |
 |
| Unusual noncrystallographic symmetry in crystals of a 420 kDa crustacean clottable protein. Kollman JM, Doolittle RF Acta Crystallographica D 2005 |
 |
| Crystal structure of native chicken fibrinogen at 2.7 Å resolution. Yang Z, Kollman JM, Pandi L, Doolittle RF Biochemistry 2001 |
 |
| Determining the relative rates of change for prokaryotic and eukaryotic proteins with anciently duplicated paralogs. Kollman JM, Doolittle RF Journal of Molecular Evolution 2000 |
 |
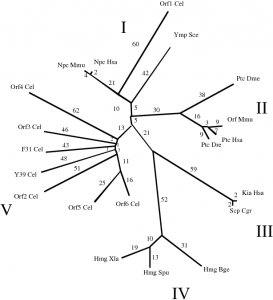
| The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH Jr. Journal of Molecular Microbiology & Biotechnology 1999 |
 |
| Is FatP a long-chain fatty acid transporter? Saier MH Jr, Kollman JM Molecular Microbiology 1999 |
 |