We generally recommend at least 600 micro Litre of solvent volume to observe un-distorted lineshapes, for small molecule NMR, where mostly we use Organic solvents that tend to show narrow lines. The lineshape of Organic solvent peak tend to closely resemble a Lorentzian curve due to its long T2. This means that it is much more prone to distortions close to the baseline, if the higher order shims are not properly adjusted. There is another condition that can exactly mirror this ‘poor higher order shim’ effect.
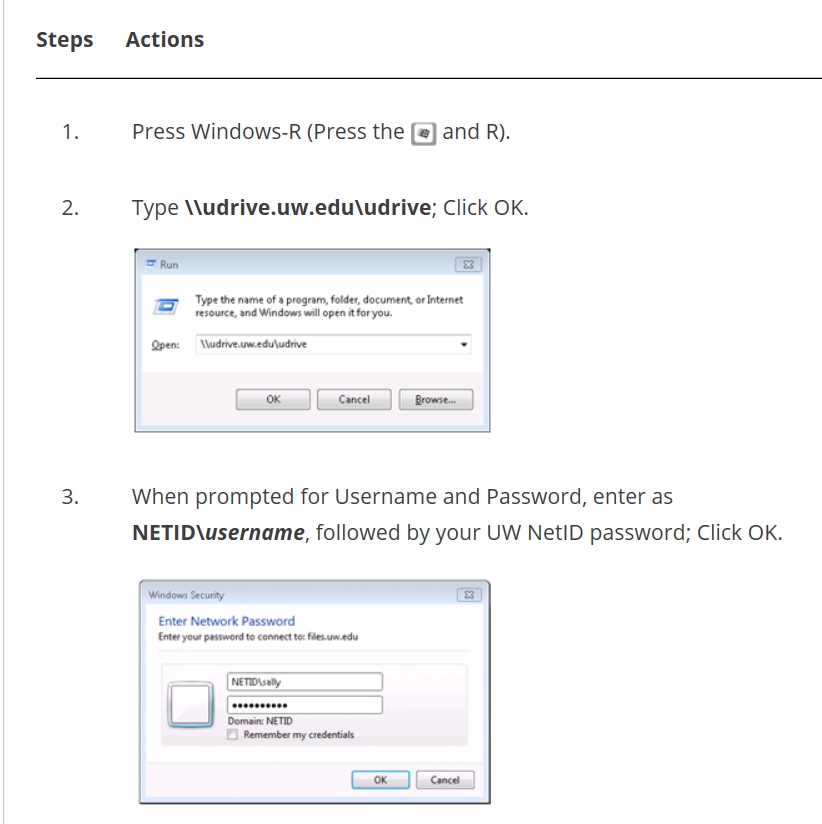
The NMR sample coil i.e. the inductance coil that picks up the NMR response after a pulse of RF is applied, is about 25 mm long.
The 1H nuclei that completely fill up a cylindrical volume of this height are the only ones that provide most of the observable NMR signal. But, limiting the sample volume that reaches only this height leads to a problem. There is an abrupt transition from the liquid volume to the air (or gas such as N2) above this column of liquid.  The magnetic susceptibility of these two materials are vastly different such that the way they ‘bend’ or distort the magnetic field lines of the static B0 field are quite different. The shim system, actually functions on the singular assumption that the sample is, ideally, an infinitely long tubular column of liquid. In other words, B0 field lines are distorted by a single medium and the distortions are smoothly continuous. Such a variation is easy to model so that the shim system can apply gradients that are of opposite signs to the inherent distortion gradients and cancel them out. The net effect is that all the protons within this volume pretty much feels the effect of only one value of B0 i.e. a highly homogeneous magnetic field.
The magnetic susceptibility of these two materials are vastly different such that the way they ‘bend’ or distort the magnetic field lines of the static B0 field are quite different. The shim system, actually functions on the singular assumption that the sample is, ideally, an infinitely long tubular column of liquid. In other words, B0 field lines are distorted by a single medium and the distortions are smoothly continuous. Such a variation is easy to model so that the shim system can apply gradients that are of opposite signs to the inherent distortion gradients and cancel them out. The net effect is that all the protons within this volume pretty much feels the effect of only one value of B0 i.e. a highly homogeneous magnetic field.
But, an abrupt jump or a ‘kink’ in the way the distortions occur along the vertical axis, makes the above method of cancelling out the existing gradients pretty difficult. The shim system and the software algorithm try their best. It is similar to fitting a very noisy raw data with as good a fitting curve as possible. Since the higher order shims are mostly responsible for optimizing the lineshape closer to the edge of the sample coil, they are the ones that get mis-set during shimming. This mis-set value of higher order shims such as Z5 and Z6, translate into an artificial assymetric lineshape. For Organic solvents that are fully deuterated, this is the predominant distortion that we have to deal with.
When you use samples that are dissolved in H2O or H2O+D2O, the rather large solvent peak at 4.7 ppm needs to be suppressed cleanly so that all the solute resonances are clearly identifiable. Generally the concentration of the H2O molecules are 3 to 4 orders or magnitude higher than that of Solutes. For this case of ‘solvent suppression’ also, the optimal shimming is very critical and an adequate amount of minimum volume becomes important.
The overlaid spectra of two cases shown below demonstrates this nicely. I recorded the red colored spectrum on our 800 MHz spectrometer with a cryoprobe. Even though the Topshim method minimized the B0 field variation to the minimum possible, the water suppression pulse sequence led to these spurious wiggle like structure that spans almost 0.7 ppm or 560 Hz. Arrow 1 points to the position of water resonance at 4.7 ppm. Positions 2 and 3 point to the peaks that are of main interest in the spectra and these are rather weak. Although the absolute intensity of water resonance (Peak 1) is effectively reduced, the wiggles that arise out of the water suppression method drowns out the peaks of importance. The volume of this sample was close to 500 micro L.
The remedy was to just add 100 microL of solvent i.e. water and the result now is dramatically improved. The green colored spectrum is the result. All the important peaks are clearly visible. I had used the same number of scans for both these spectra and the dynamic range of the green spectra is much better than the former.

Conclusion When you have limited availability of solute, choosing the correct volume of solvent to run NMR, such that you maximize the concentration and get ‘all’ the Nuclei give useful signal, is a balancing act.
HAPPY SPINNING !