Research
The Bomsztyk lab has had a long-standing interest to understand disease pathways and develop a variety of tools to reverse-engineer the molecular processes of disease.
Early on, the lab focused on ions and developing tools and methods to study ion fluxes, including first-of-its-kind microelectrodes to measure ionized calcium, pH, pCO2, and HCO3– in tissues and bodily fluids. This was followed by the development of the microelectrometric method to measure intracellular chloride and atomic absorption spectroscopy to measure intracellular sodium and potassium. Later on, the lab focused on signal transduction of nuclear events and the identification and cloning of proteins and how they interact to control transcription and epigenetic processes.
With the recognition that these processes are complex, over the last fifteen years the lab has been focused on developing fast, high-throughput and user-friendly methods, devices, and software to study transcription and epigenetic processes. This includes the Fast-ChIP, Matrix-ChIP, Matrix-MeDIP, Matrix-RT platforms, as well as the GraphGrid/PCRCrunch software to acquire, analyze, and visualize PCR data generated by these platforms. These tools have been used in more than a thousand of publications.
To match the power and throughput of these analytical tools, the Bomsztyk Lab, in collaboration with the Tom Matula Lab at UW Applied Physics Laboratory, engineered and built a high-throughput microplate sample preparation instrument, PIXUL. This multipurpose sample preparation instrument is being used in high-throughput genomic, epigenetic, and proteomic applications.
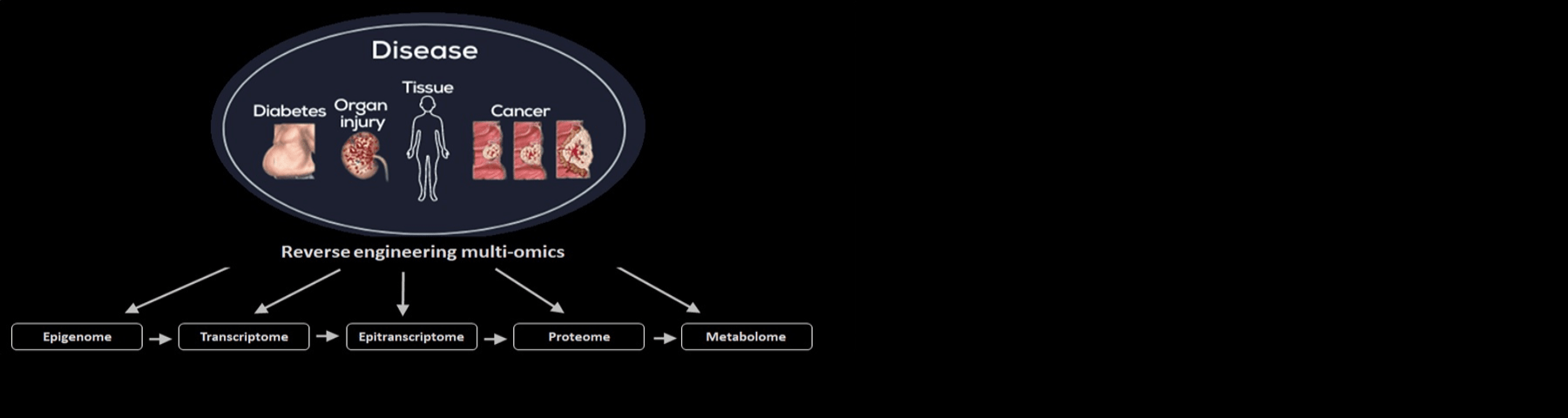
With the focus on disease, the Bomsztyk Lab continues to advance a host of user-friendly high-throughput omics methods/devices and to integrate these transcription/epigenetic technologies with next generation sequencing (NGS). These platforms are now employed to interrogate tissues and organs, including FFPEs, to reverse-engineer pathways involved in organ injury, cancer, and diabetes as a way to discover disease biomarkers and drug targets.